Home /
Expert Answers /
Computer Science /
kindly-code-in-nbsp-simple-python-language-task-2-virus-mutation-simulator-pa356
(Solved): Kindly code in simple python language. Task 2 - Virus mutation simulator ...
Kindly code in simple python language.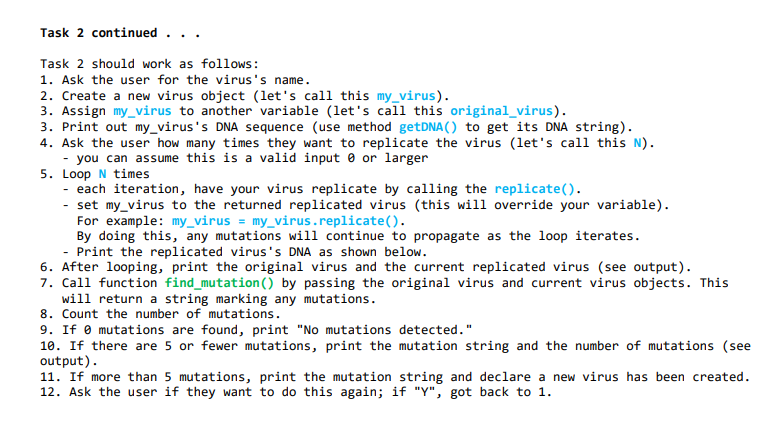
Task 2 - Virus mutation simulator Task 2 will emulate a simple virus. The virus is represented by a DNA sequence. Recall that DNA is composed of only four kinds of nuclear acids represented by the leters: A, G, C, T. Our DNA sequence will be a string of 50 DNA letters. Each time we replicate the virus, there is a \( 5 \% \) chance our DNA will mutate. If more than 5 mutations occur (i.e., 10\% of our starting DNA), we will claim a new virus has emerged. For this task, a class named virus has already been defined for you in trinket. Class name: virus Instance data (1) DNA A 50 characters string containing only the letters ' \( A \) ', ' \( \mathrm{G}^{\prime} \), ' \( T \) ', ' \( C \) '. Methods (3) _init__(DNAinput="") DNAinput is a string, with default value "" (empty string). param: DNAinput is a string. - If DNAinput is passed to when creating a virus object, assign DNAinput to self.DNA - You should check to ensure that the DNAinput string is exactly 50 characters and is only from the valid character set. (For this task, this should always be true, but it is good practice to check.) - If the DNAinput string is empty, then your method should randomly generate a DNA sequence of 50 characters, where each character is only 'A', ' \( G \) ', ' \( T \) ', ' \( C \) '. Assign the random DNA to self. DNA. getDNA() No parameters, returns a string - This is a "getter" method that returns the value self.DNA
replicate() No parameters, returns a virus object! <- pay attention. The return is a new object - This method will create a new virus object based on the DNA of this virus object. - However, before passing this virus's DNA string, do the following to check if there is a mutation: Generate a random number between 1-100. If the random number is larger than 94, mutate the DNA. How to mutate the string? Randomly select a number between 0-49. Now, change the DNA value at that position to a randomly selected ' \( A \) ', ' \( G \) ', ' \( T \) ', or ' \( C \) '. (This simulates a mutation in the DNA.) - If there is a mutation, create a new virus with the mutated string, for example: \( x= \) virus(mutatedDNA) - Otherwise, create a new virus using the original virus DNA, for example: \( x=v i r u s \) (self.DNA) - Return the created virus object (e.g., x) You also need to write the following function: find_mutation(virus1, virus2) \( \quad \rightarrow \) returns a string - This function (not an object method, but a stand-alone function) takes two virus objects and returns a string that marks the changes between the two viruses' DNAs. How? - Get each object's DNA string (use the getDNA() method) - Loop through the string and check where virus1 and virus2 string characters are different. - As you loop through the strings, create a new string with a "^" when a difference is encountered, and a space " "otherwise. - For example: Virus 1 DNA is: "AACTATCGCGAATAATCGTAAAAGCCCAGCATGCCAGGTGCGGTATCTGG" Virus 2 DNA is: "AACTATCTCGAATAATCGTAAAGACCCTACATGCCAGGTGCGGTATCTGG" Your string :" \( \wedge \) " \( \wedge \wedge \) mutation mutations Return the string indicating the mutations. The number of "\( { }^{\wedge} \) " in the string is the number of mutations that occurred.
Task 2 continued ... Task 2 should work as follows: 1. Ask the user for the virus's name. 2. Create a new virus object (let's call this my_virus). 3. Assign my_virus to another variable (let's call this original_virus). 3. Print out my_virus's DNA sequence (use method getDNA() to get its DNA string). 4. Ask the user how many times they want to replicate the virus (let's call this N). - you can assume this is a valid input 0 or larger 5. Loop \( N \) times - each iteration, have your virus replicate by calling the replicate(). - set my_virus to the returned replicated virus (this will override your variable). For example: my_virus = my_virus. replicate(). By doing this, any mutations will continue to propagate as the loop iterates. - Print the replicated virus's DNA as shown below. 6. After looping, print the original virus and the current replicated virus (see output). 7. Call function find_mutation() by passing the original virus and current virus objects. This will return a string marking any mutations. 8. Count the number of mutations. 9. If 0 mutations are found, print "No mutations detected." 10. If there are 5 or fewer mutations, print the mutation string and the number of mutations (see output). 11. If more than 5 mutations, print the mutation string and declare a new virus has been created. 12. Ask the user if they want to do this again; if "Y", got back to 1 .
Expert Answer
COde with required class and methods: import random as r #class virus class virus: def __init__(self,DNAinput=""): self.DNA=DNAinput def getDNA(self):